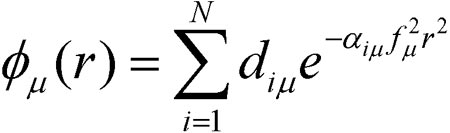
S-Type Basis Function
A set of standard basis sets is stored internally in Gaussian (see the Basis Sets section earlier in this chapter); these basis sets may be specified by including the appropriate keyword within the route section for the calculation. The Gen keyword allows a user-specified basis set to be used in a Gaussian calculation. It is used in the place of a basis set keyword or a density fitting basis set keyword. In this case, the basis set description must be provided as input (in a separate basis set input section).
Gen may be used in a completely analogous way to specify an alternate density fitting basis set (see the examples).
The GenECP variation may be used to read in both basis functions and ECPs; it is equivalent to Gen Pseudo=Read. It is designed for use in ONIOM calculations in which you want to use a general basis set with ECPs within one ONIOM layer.
The GFPrint keyword may be used to include the gaussian function table within the output file. The GFInput keyword may be used to have the table printed in a form which is suitable for input to Gen. The ExtraBasis keyword may be used to make additions to standard basis sets. Similarly, the ExtraDensityBasis keyword may be used to make additions to standard density fitting basis sets.
A brief general overview of basis functions is provided as the final subsection of this discussion.
External basis sets are read into Gaussian by specifying Gen (for general basis) in the route section. The keywords 5D, 6D, 7F, and 10F are used to specify use of Cartesian or pure d and f (and higher) functions; the defaults are 5D and 7F. All d-shells in a calculation must have the same number of functions. Similarly, f- and higher shells must either be all Cartesian or all pure.
Defining a shell. External basis input is handled by the routine GenBas in Link 301. The basic unit of information that it reads from the basis set input section is the shell definition block. A shell definition block, together with the global specification of pure vs. Cartesian functions, contains all necessary information to define a shell of functions. It consists of a shell descriptor line, and one or more primitive gaussian lines:
| IType | NGauss Sc | Shell descriptor line: shell type, # primitive gaussians, and scale factor. |
| α1 | d1μ | Primitive gaussian specification: exponent and contraction coefficient. |
| α2 | d2μ | |
| … | ||
| αN | dNμ | There are a total of NGauss primitive gaussian lines. |
IType defines the shell type and shell constraint and may be S, P, D, SP, SPD, F, G, …, for an s-shell, p-shell, d-shell, sp-shell, f-shell, g-shell, and so on. NGauss specifies the number of primitive gaussian shells (the degree of contraction) for the shell being defined. The shell scale factor is given by Sc (i.e., all primitive exponents are scaled by Sc2).
The subsequent NGauss primitive gaussian lines define the exponents αk and contraction coefficients, dkμ. Each line provides the exponent for one primitive, followed by its contraction coefficient (or s and p coefficients for an sp-shell).
A second format also exists to specify a shell as a least-squares gaussian expansion of a Slater orbital. This is requested by a shell descriptor line of the form STO, IOrb, NGauss, Sc. IOrb is one of 1S, 2S, 2P, 2SP, 3S, 3P, 3SP, 3D, 4SP, and specifies which expansion is requested. Note that 2SP requests the best least-squares fit simultaneously to S and P slater orbitals and is not equivalent to separately specifying the best S and the best P expansions. NGauss is the same as above. Gaussian expansions of Slater functions having from 1 to 6 primitives are available. Sc is the scale factor and hence the exponent of the slater function being expanded. No primitive gaussian lines are required after a shell descriptor line requesting an STO expansion.
Defining the basis for an atom or atom type. One customarily places at least one, and often several, shells on any given nuclear center (“atom”), via a center definition block. A center definition block consists of a center identifier line, and one shell definition block for each shell desired on the center(s) specified. It is terminated by a line with either asterisks or plus signs in columns 1 through 4:
| c1 c2 … 0 | Center identifier line: specifies applicability for these shells. | |
| IType | NGauss Sc | First shell definition block. |
| α2 | d2μ | |
| … | ||
| αN | dNμ | |
| … | Additional shell definition blocks. | |
| IType | NGauss Sc | Final shell definition block. |
| α2 | d2μ | |
| … | ||
| αN | dNμ | |
| **** | Separator: terminates the center definition block. | |
The center identifier line specifies a list of centers on which to place the basis functions in the center definition block, terminated by a 0. It can contain one or more integers, which are used to indicate the corresponding atom(s) in the molecule specification; more commonly, it contains a list of atomic symbols to refer to all atoms of a specific type. Center numbers and atomic symbols may be freely intermixed within a single center identifier line.
To help detect input mistakes, if a center definition block specifies an atom that is not present in the molecule, the run is aborted. If the center is preceded by a minus sign (e.g. -H), the basis set information is simply skipped if no atom of that type is present in the molecule specification (the terminal zero may also be omitted in this case). The latter syntax is intended for creating basis set include files that specify a standard basis set for many atoms; once built, it can be included in its entirety in the input stream when the basis set is desired, via the include (@) function (as described earlier in this chapter).
A center or atom type may be specified in more than one center definition block. For example, in the Gaussian 09 basis set directory—$g09root/g09/basis on UNIX systems—there is one file which specifies 6-31G as a general basis set (631.gbs), and another file containing d exponents which would be included as well to specify 6-31G* (631s.gbs). Every atom from H through Cl is specified in both files, and in practice both of them would be included (most often along with additional basis set specifications for those atoms in the molecule for which the 6-31G basis set is not available).
Basis set transformation option to the Integral keyword. Several options to the Integral keyword control whether/how generalized contraction basis sets are transformed to reduce the number of primitives. Int=BasisTransform=N says to transform generalized contraction basis sets to reduce the number of primitives, neglecting primitives with coefficients of 10-N or less. This is the default, with N=4. Int=ExactBasisTransform says to transform generalized contraction basis sets to reduce the number of primitives, but using only transformations which are exact (do not change the computed energy). Finally, Int=NoBasisTransform says not to transform generalized contraction basis sets to reduce the number of primitives.
Drawing on Pre-Defined Basis Sets in Gen Input. Gaussian adds flexibility to general basis set input by allowing them to include pre-defined basis sets within them. Within a center definition block for an atom type (or types), an entire shell definition block may be replaced by a line containing the standard keyword for a pre-defined basis set. In this case, all of the functions within the specified basis set corresponding to the specified atom type(s) will be used for all such atoms within the molecule.
The SDD, SHF, SDF, MHF, MDF, MWB forms may be used to specify Stuttgart/Dresden basis sets/potentials within Gen basis input. Note that the number of core electrons must be specified.
Here is a portion of the Gen input corresponding to the 6-31+G(d) basis set:
H 0 Applies to all hydrogen atoms. S 3 1.00 0.1873113696D+02 0.3349460434D-01 0.2825394365D+01 0.2347269535D+00 0.6401216923D+00 0.8137573262D+00 S 1 1.00 0.1612777588D+00 0.1000000000D+01 **** C 0 Applies to all carbons. S 6 1.00 6-31G functions. 0.3047524880D+04 0.1834737130D-02 0.4573695180D+03 0.1403732280D-01 0.1039486850D+03 0.6884262220D-01 0.2921015530D+02 0.2321844430D+00 0.9286662960D+01 0.4679413480D+00 0.3163926960D+01 0.3623119850D+00 SP 3 1.00 0.7868272350D+01 -0.1193324200D+00 0.6899906660D-01 0.1881288540D+01 -0.1608541520D+00 0.3164239610D+00 0.5442492580D+00 0.1143456440D+01 0.7443082910D+00 SP 1 1.00 0.1687144782D+00 0.1000000000D+01 0.1000000000D+01 D 1 1.00 Polarization function. 0.8000000000D+00 0.1000000000D+01 **** C 0 Applies to all carbons. SP 1 1.00 Diffuse function. 0.4380000000D-01 0.1000000000D+01 0.1000000000D+01 ****
The following Gen input uses the 6-31G(d,p) basis set for the carbon and hydrogen atoms and the 6-31G‡ basis set for the fluorine atoms in the molecule, and places an extra function only on center number 1 (which happens to be the first carbon atom in the molecule specification for 1,1-difluoroethylene):
C H 0 6-31G(d,p) **** F 0 6-31G(d',p') **** 1 0 Place a diffuse function on just one carbon atom. SP 1 1.00 0.4380000000D-01 0.1000000000D+01 0.1000000000D+01 ****
The following job uses the Gaussian include file mechanism to specify the basis functions for chromium:
# Becke3LYP/Gen Opt Test HF/6-31G(*) Opt of Cr(CO)6 molecule specification C O 0 6-31G(d) **** @/home/gwtrucks/basis/chrome.gbs/N
Note that .gbs is the conventional extension for basis set files (for gaussian basis set).
The following example uses general basis set input to specify both the basis set and the density fitting basis set.
# RBLYP/GEN/GEN 6D
HCl: reading in 6-31g* AO basis and DGA1 fitting set.
6D is specified because the default for general basis
input is 5D but the 6-31g* basis is defined to use 6D
0,1
cl
h,1,1.29
! here are the 6-31g* basis sets for Cl and H
cl 0
S 6 1.00
0.2518010000D+05 0.1832959848D-02
0.3780350000D+04 0.1403419883D-01
0.8604740000D+03 0.6909739426D-01
0.2421450000D+03 0.2374519803D+00
0.7733490000D+02 0.4830339599D+00
0.2624700000D+02 0.3398559718D+00
SP 6 1.00
0.4917650000D+03 -0.2297391417D-02 0.3989400879D-02
0.1169840000D+03 -0.3071371894D-01 0.3031770668D-01
0.3741530000D+02 -0.1125280694D+00 0.1298800286D+00
0.1378340000D+02 0.4501632776D-01 0.3279510723D+00
0.5452150000D+01 0.5893533634D+00 0.4535271000D+00
0.2225880000D+01 0.4652062868D+00 0.2521540556D+00
SP 3 1.00
0.3186490000D+01 -0.2518280280D+00 -0.1429931472D-01
0.1144270000D+01 0.6158925141D-01 0.3235723331D+00
0.4203770000D+00 0.1060184328D+01 0.7435077653D+00
SP 1 1.00
0.1426570000D+00 0.1000000000D+01 0.1000000000D+01
D 1 1.00
0.7500000000D+00 0.1000000000D+01
****
h 0
S 3 1.00
0.1873113696D+02 0.3349460434D-01
0.2825394365D+01 0.2347269535D+00
0.6401216923D+00 0.8137573261D+00
S 1 1.00
0.1612777588D+00 0.1000000000D+01
****
! here are the DGA1 fitting sets for Cl and H
cl 0
S 1 1.00
0.2048000000D+05 0.1000000000D+01
S 1 1.00
0.4096000000D+04 0.1000000000D+01
S 1 1.00
0.1024000000D+04 0.1000000000D+01
S 1 1.00
0.2560000000D+03 0.1000000000D+01
S 1 1.00
0.6400000000D+02 0.1000000000D+01
SPD 1 1.00
0.2000000000D+02 0.1000000000D+01 0.1000000000D+01 0.1000000000D+01
SPD 1 1.00
0.4000000000D+01 0.1000000000D+01 0.1000000000D+01 0.1000000000D+01
SPD 1 1.00
0.1000000000D+01 0.1000000000D+01 0.1000000000D+01 0.1000000000D+01
SPD 1 1.00
0.2500000000D+00 0.1000000000D+01 0.1000000000D+01 0.1000000000D+01
000000D+01 0.1000000000D+01
****
h 0
S 1 1.00
0.4500000000D+02 0.1000000000D+01
S 1 1.00
0.7500000000D+01 0.1000000000D+01
S 1 1.00
0.1500000000D+01 0.1000000000D+01
S 1 1.00
0.3000000000D+00 0.1000000000D+01
****
If you wanted to specify the density fitting basis set with general basis set input, then you would use a route section like this one (substituting the appropriate basis set for your problem):
# RBLYP/6-31G(d,p)/Gen 6D
ExtraBasis, ExtraDensityBasis, GFInput, GFPrint, Integral, Pseudo
A single basis function is composed of one or more primitive gaussian functions. For example, an s-type basis function φμ(r) is:
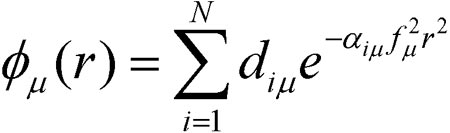
S-Type Basis Function
N is the number of primitive functions composing the basis function, and it is called the degree-of-contraction of the basis function. The coefficients diμ are called contraction coefficients. The quantities αiμ are the exponents, and f is the scale factor for the basis function. The maximum degree-of-contraction permitted in Gaussian is 100.
A shell is a set of basis functions φμ with shared exponents. Gaussian supports shells of arbitrary angular momentum: s, p, d, f, g, h, and so on. An s-shell contains a single s-type basis function. A p-shell contains the three basis functions pX, pY, and pZ. An sp-shell contains four basis functions with common gaussian exponents: one s-type function and the three p-functions pX, pY and pZ.
A d-shell may be defined to contain either the six second-order functions (dX2, dY2, dZ2, dXY, dXZ, dYZ), or the five so-called “pure d” basis functions (d z2-r2, dx2-y2, dxy, dxz, dyz). Likewise, an f-shell may contain either the 10 third-order gaussians or the 7 “pure f” functions. Higher order shells function similarly. Note that the contraction coefficients in a shell must be the same for all functions of a given angular momentum, but that s and p contraction coefficients can be different in an sp-shell. A scale factor is also defined for each shell. It is used to scale all the exponents of primitives in the shell. The program has the ability to convert between the two types of functions [Schlegel95a].
Consider the series of basis sets STO-3G, 6-31G, and 6-311G(d) for the carbon atom. With the STO-3G, basis there are two shells on a carbon atom. One is an s-shell composed of 3 primitive gaussian functions (which are least-squares fit to a Slater 1s orbital). The other sp-shell is a least-squares fit of 3 gaussians to Slater 2s and 2p orbitals with the constraint that the s and p functions have equal exponents. These expansions are the same for all atoms. Only the scale factors for each shell differ from atom to atom. For carbon atoms, the 1s- and 2sp-shells have scale factors of 5.67 and 1.72, respectively. The 6-31G basis on a first row atom has three shells. One shell is a contraction of six primitive s-type gaussians. The second shell is a combination of three primitive sp-shells. The third shell consists of a single sp-function. These functions were optimized for the atom. Scale factors of 1.00, 1.00, and 1.04, respectively, for each shell for carbon were then determined by molecular calculations. As its name implies, the 6-311G(d) basis has 5 shells: an s-shell with 6 primitives, 3 sp-shells with 3, 1, and 1 primitives, and an uncontracted d-shell. All shells are unscaled (have unit scale factor).
Last updated on: 10 May 2009